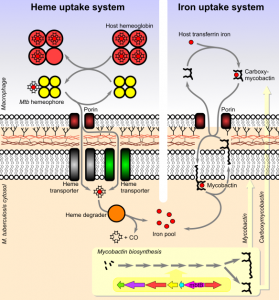
Mycobacterium tuberculosis Iron/Heme Uptake Pathways
Iron is an essential metal for life. Mycobacteria must import iron from its host. Molecules involved in iron chelating pathways are well characterized, such as those involving siderophores. Recently, it has been shown that during the early stages of S. aureus infection the major source of nutrient iron is heme rather than transferrin iron. A potential mycobacterial secreted hemophore has been identified in mycobacteria. Also, a potential cytosolic heme-degrading protein, MhuD, has been identified. Finally, two potential heme transporters have been identified in playing a role in heme-uptake. Hence, a novel mcobacterial heme uptake system may exists. My laboratory will be dissecting this pathway both structrually and biochemically. To complement the heme uptake pathway, we are also studying the mechanism of siderophore-dependent iron uptake. Investigations into these uptake pathways are on-going.
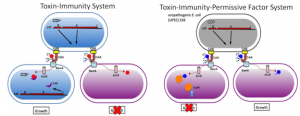
Contact Dependent Growth Inhibition
Contact-dependent growth inhibition (CDI) systems are present in a wide variety of Gram-negative bacteria, including many important pathogens, where they appear to function in intra-species growth competition and communication. CDI+ cells physically interact with susceptible bacteria and deliver a protein toxin that inhibits target cell growth. Inhibition is mediated by CdiA, a large filamentous protein that is expressed on the surface of inhibitor cells. The C-terminal region of CdiA (CdiA-CT) is polymorphic and contains the growth inhibition activity, suggesting that CDI systems deploy a variety of different toxins. CDI loci also encode highly variable CdiI immunity proteins, which specifically bind cognate CdiA-CT toxins to prevent autoinhibition (left panel). A survey of available bacterial genomes revealed approximately 120 distinct families of CdiA-CT/CdiI toxin/immunity pairs. Moreover, we have recently discovered that some CdiA-CT domains interact with specific target cell proteins termed permissive factors, and these binding interactions are required to activate the delivered toxins (right panel). Thus, some CdiA-CT domains interact with both their cognate CdiI immunity protein and a target cell permissive factor. Our long-term goal is to leverage the knowledge gained from this work to develop novel and targeted antimicrobial therapies. In this proposal, we focus on the structural, functional and genetic analyses of CdiA-CT toxins and their interacting protein partners. We will elucidate the mechanisms by which CdiA-CT toxins are neutralized by CdiI proteins, and activated by permissive factors, thereby yielding insights into the intricate toxin-immunity network encoded by bacterial CDI systems.