Neuroimaging Data Model (NIDM)
 The Neuroimaging Data Model (NIDM) is a collection of specification documents that define extensions the W3C PROV standard for the domain of human brain mapping. NIDM uses provenance information as means to link components from different stages of the scientific research process from dataset descriptors and computational workflow, to derived data and publication. For more information on our recent NIDM-Terms grant (1RF1MH120021-01) see NIDM-Terms
The Neuroimaging Data Model (NIDM) is a collection of specification documents that define extensions the W3C PROV standard for the domain of human brain mapping. NIDM uses provenance information as means to link components from different stages of the scientific research process from dataset descriptors and computational workflow, to derived data and publication. For more information on our recent NIDM-Terms grant (1RF1MH120021-01) see NIDM-Terms
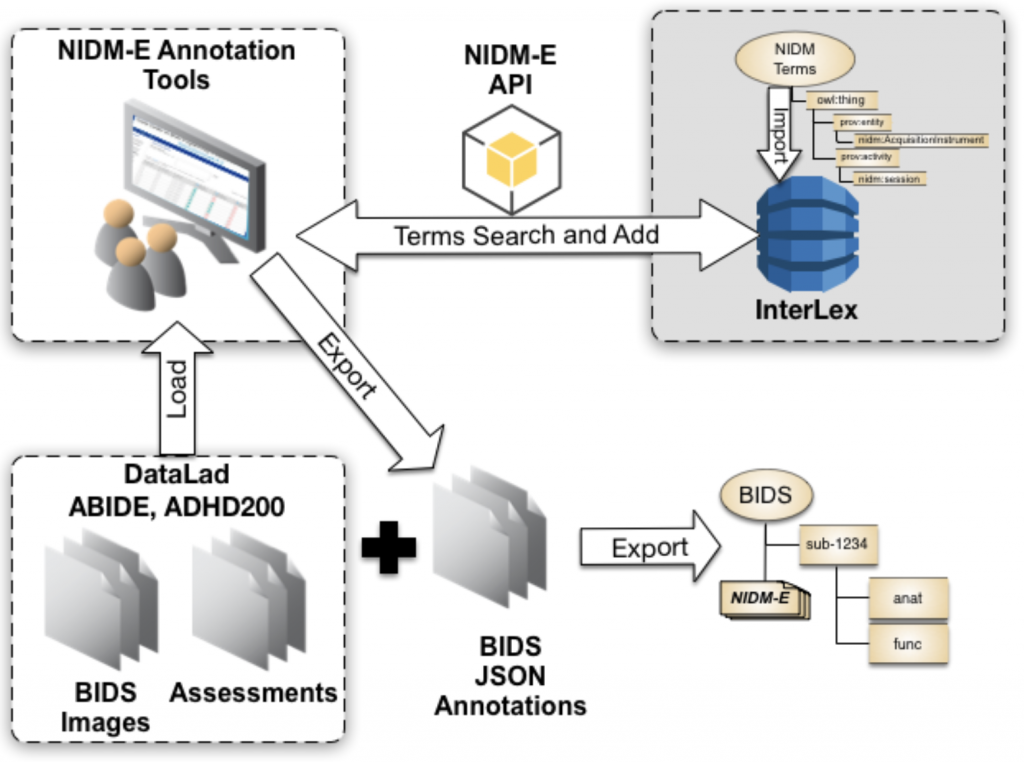
PyNIDM is a python3-based application programmer’s interface (API) with various tools for working with NIDM documents. PyNIDM contains a hierarchical class library consistent with the project-session-acquisition hierarchy used in NIDM-Experiment documents (see http://nidm.nidash.org/specs/nidm-experiment_dev.html). The API also contains query functions that developers can use to query NIDM documents and have been incorporated into the pynidm query tool. Additional tools include `bidsmri2nidm` which exports metadata from BIDS datasets and creates a NIDM-Experiment metadata file along with providing support for annotating BIDS data, creating data element definitions referred to as “BIDS sidecar files” and provides the user with the ability to link specific data elements to broader concepts useful for high-level queries across datasets (e.g. find all datasets with a measures of intelligence, age, and handedness regardless of how those were measured and with what units). These annotation activities are critical in making BIDS datasets more interoperable and findable . For annotating CSV-based phenotype information, pynidm includes a `csv2nidm` tool which, similar to the bids tools, allows one to annotate csv files and save the resulting data as a NIDM-Experiment file. The tool supports adding to existing NIDM files therefore providing a suite of tools to create NIDM representations of imaging datasets and associated phenotype information. When combined with NIDM-supported tools for adding brain volume derivative data (see freesufer, fsl, and ants tools), we provide a suite of tools for capturing semantically-annotated data and metadata in NIDM for use in broad-based query across datasets including derived data.
Templates for Brain Segmentation in Down Syndrome
In this study, we create disease-specific brain atlases for Down Syndrome (DS) research and quantify the accuracy of using these atlases as compared with using an individualized MRI-based atlas to calculate amyloid burden in brain regions implicated in Alzheimer’s Disease (AD) progression. Regional PET amyloid burden assessments typically use an individualized Magnetic Resonance Imaging (MRI)-based brain segmentation (i.e. atlas) to identify anatomical boundaries in brain regions. When participants cannot tolerate an MRI, a standardized brain atlas is used, often constructed from a single, young, healthy individual, whose brain does not reflect the amount of brain atrophy typically seen in the progression of AD. For this reason, Disease-specific high-resolution brain atlases were created by joint label fusion of individualized segmentations (labels) computed from each participant’s T1-weighted MRI in a leave-one-out (LOO) cross-validation paradigm. The total sample included participants with consensus diagnoses of non-demented (n=55), mild cognitive impairment (n=19), and dementia (n=11). Amyloid PET scans for each participant were registered to the appropriate LOO disease-specific atlas, and the average amyloid burden was compared to the ground-truth of using individualized MRI-based atlas. We hypothesize that there will be high correlation between using standardized atlases and individualized MRI-based atlases. Further, knowing a participant’s diagnosis and using an appropriate standardized disease-specific atlas will hold the highest correlations with individualized MRI-based atlases and likely be more accurate relative to the ground-truth than using a standard atlas constructed from either a different diagnostic group or from all DS participants of similar age.
Neuroimaging Biomarkers of Dementia in Down Syndrome
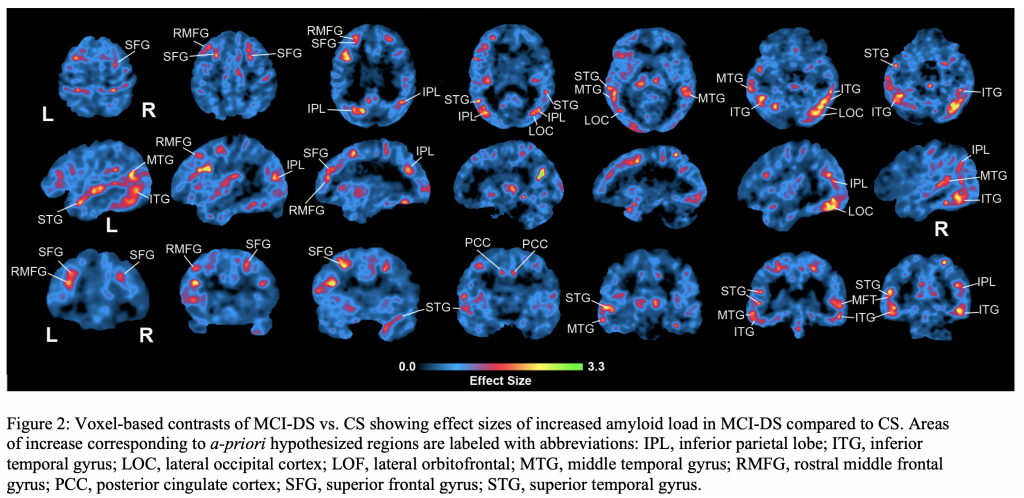
Down Syndrome (DS) is associated with higher risk of dementia. We hypothesize amyloid-β (Aβ) in specific brain regions differentiate mild cognitive impairment (MCI-DS) in DS and test these hypotheses using cross-sectional and longitudinal data. 18F-AV-45 positron emission tomography (PET) data was collected to analyze amyloid burden in 58 participants clinically classified as cognitively stable (CS) or MCI-DS and 12 longitudinal CS participants. The study confirmed our hypotheses of increased amyloid in inferior parietal, lateral occipital, and superior frontal regions as the main effects differentiating MCI-DS from the CS groups. The largest annualized amyloid increases in longitudinal CS data were in rostral middle frontal, superior frontal, superior/middle temporal, and posterior cingulate cortices. This study helps us understand amyloid in the MCI-DS transitional state between cognitively stable aging and frank dementia in DS. The spatial distribution of Aβ may be reliable indicators of MCI-DS in DS.