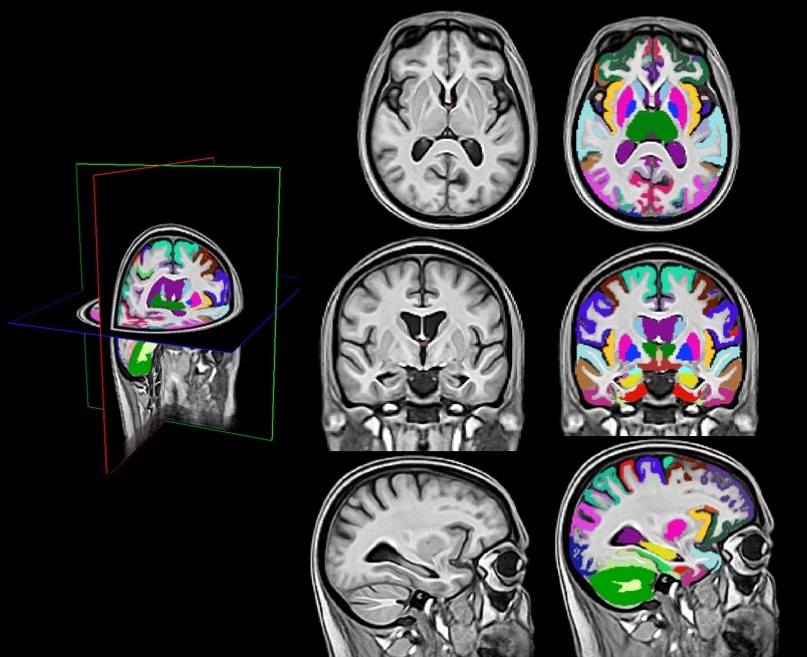
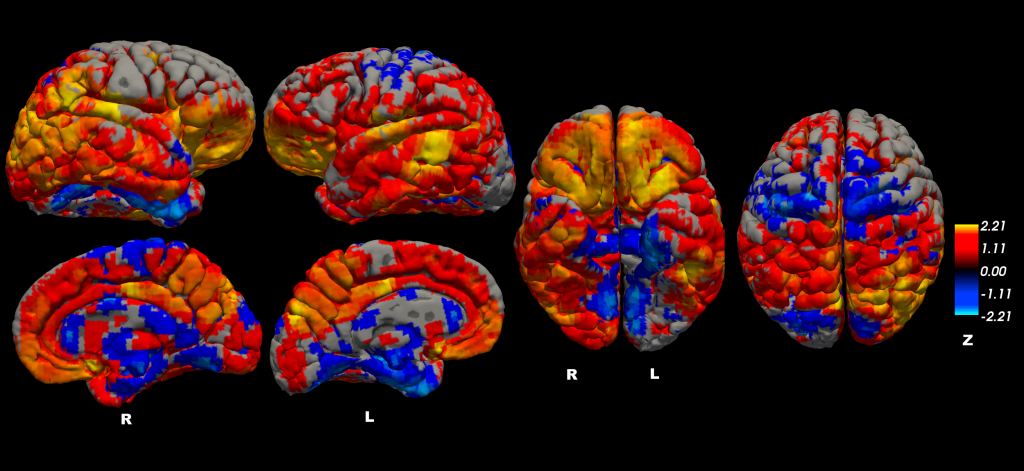
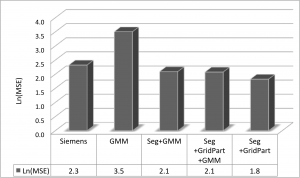
Congratulations to undergraduate student Nazek Queder who was awarded a research stipend to support her work on “Creating a New Brain Template for PET Studies of Alzheimer’s disease in the Down Syndrome Population,” under the supervision of Dr. David Keator. Nazek will work on state-of-the art templates to improve our ability to understand regional amyloid accumulation in participants who are unable to tolerate an MRI scan.
Nazek is a 4th year psychology student with a broad experience in psychology, neuroscience, art, and programming. Nazek’s passion is to help us understand how the brain works and is “thrilled to be able to contribute to society by us having more tangible measures to read and model brain atrophy in patients with Alzheimer’s Disease.”