Michael McClelland’s lab works in the area of genomics, a field that aims to interpret the language of DNA.
Salmonella genomics: Dr McClelland organizes the sequencing of major pathogens, including Salmonella enterica sv Typhimurium, resulting in papers in Nature and Nature Genetics. The lab also provides resources and techniques to over 50 labs worldwide for the study of Salmonella genomes, transcription, and mutant library screens. Among those resources are high-complexity genome-wide random and ordered mutant collections, representing eight clinically important Salmonella serovars, and E. coli, as illustrated below.
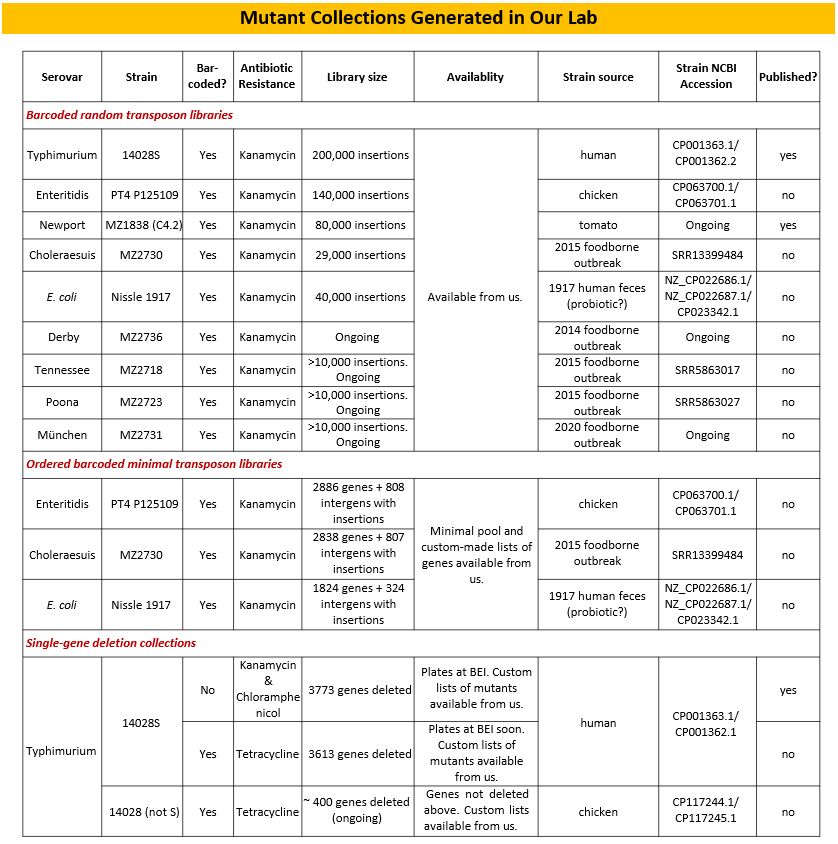
We continue to develop these resources for additional Salmonella serovars, including Gallinarum and Typhi.
Cancer therapy: Harmless variants of Salmonella prefer to reside in tumors over any other location in the human body, by a factor of 1000 or more. These bacteria can cause cancer cures. The lab has engineered improvements in Salmonella as a therapeutic and novel delivery agent in cancer. Newly developed high-throughput tools are used to identify promoters that are turned on only in tumors. Using such specific promoters, cloned foreign therapeutic proteins could be produced only when bacteria reach tumors. In addition, tools have been developed to track thousands of bacterial mutants simultaneously, which allows mutants with favorable characteristics, such as those that accumulate even better in tumors, to be isolated and further engineered.
Prostate cancer prognostics: The McClelland lab is involved in a large multicenter grant to identify gene expression changes in prostate cancer that are associated with an increased risk of recurrence of the disease after prostatectomy. The McClelland lab performs the RNA work and much of the bioinformatics for this project. Genomics: Michael McClelland developed the first DNA cleavage methods for making physical maps of DNA molecules millions of bases long.
DNA methylation: Michael McClelland was the first to describe that the regulatory regions of mammalian genes, called promoters, contain sequences called “CpG islands”. Methylation of these features (addition of a methyl group to the DNA) is important in marking genes for regulation. Dr McClelland studies the alteration of methylation during the dysregulation of genes that occurs in cancer. As part of this work, the McClelland lab constructed the first “promoter microarrays” of DNA fragments from 10,000 promoters and collaborated on the first Chromatin immunoprecipitation-array (ChIP-chip) experiments, which identify proteins bound to promoters in living cells.
Our laboratory applies high-throughput methods in all of these areas.
Program 1: Salmonella genomics.
Background. Salmonella is among the most important orally acquired pathogens in the world. Approximately one million deaths and 100 million human infections are caused annually by these organisms, and they are major pathogens of domestic livestock. On a genomic level, strains within this species can differ by hundreds of their ~4,500 genes. This variability is the result of a mosaic of lateral transfer events within a constant genome scaffold. These differences result in an extraordinary diversity of host ranges and pathogenic presentations between strains. Known virulence functions are encoded primarily within these laterally transferred regions. However, despite 30+ years of intensive study, the functions of most genes within these important regions are still unknown. We are in the process of rectifying this gap in knowledge.
Objective. Understand the evolution of pathogenesis in Salmonella with the objective of generating principles applicable to other diseases, allowing new methods of treatment, and also exploitation of avirulent strains for human needs.
Approach. We have sequenced strains used in laboratory research and are now in the finishing stages of sequencing 300 more strains that capture the diversity of the species. We are applying and developing novel analysis tools and pipelines for high-throughput sequencing data to harvest the millions of sequence reads and convert these to knowledge about how Salmonella evolved to become a successful pathogen. Understanding sequence diversity:
- allows steps in evolution to be better understood,
- allows the development of DNA typing methods that correlate better with infectious manifestations,
- allows the construction of specific in-frame knockouts of every gene as a critical resource for our functional genomic studies.
In addition, we have created high-complexity random transposon integration mutant libraries in eight Salmonella enterica serovars (Typhimurium, Enteritidis, Newport, Choleraesuis, Derby, Tennessee, Muenchen, and Poona). We have also built barcoded and non-barcoded collections of defined single-gene and multi-gene deletion mutants in Salmonella enterica sv Typhimurium 14028S. These deletion mutants include deletions in every non-essential gene in the 14028 genome. We are using these resources to improve our knowledge of gene function, by submitting the collection(s) to various selective environments and estimating the role of each gene for the survival of the bacterium in those environments.
Program 2: Salmonella as a therapeutic delivery agent in cancer.
Background. Harmless live vaccine strains of Salmonella naturally accumulate 1000X in tumors. Tumors have lower than normal levels of oxygen, where Salmonella will continue to thrive protected from the immune system. Salmonella may also further deplete oxygen to levels thereby killing and growing on the remains of tumor cells. This leads to cures.
Objective. Improve upon the natural ability of Salmonella to kill tumors. In addition, Salmonella can be used to deliver therapeutic agents, such as enzymes for drug metabolism and cytokines.
Approaches. In order to understand and improve the ability of Salmonella to kill tumors, and develop this ability for therapeutic use we are taking the following steps:
- identify promoters active in tumors to express therapeutics,
- identify mutants that grow better than wild-type bacteria in tumors,
- identify avirulent strains that grow well in cancer,
- determine the usefulness of these mutations in a variety of animals so they can be used to treat tumors in companion animals.
Program 3: Biomarkers for prostate cancer recurrence after prostatectomy.
Background. Prostate cancer is the most common cancer in men (70% of 70-year-olds!). Tens of thousands die each year because the tumor has metastasized before treatment. However, the vast majority of prostate cancers do not lead to death even if untreated but most cases are treated, nevertheless, because there is no test for accurate identification of indolent versus aggressive cases. More than 100,000 men are treated unnecessarily each year in the US, at a cost of billions of dollars, and with some side-effects in most cases.
Objective. Develop prognostic tests to accurately define those patients with progressive disease, so they can be targeted for early aggressive treatment.
Approaches. We undertook the following steps towards our goal:
- accumulate clinical data over decades for hundreds of consented patients,
- develop a method that assigns predictive markers to tumor or adjacent reactive tissues (Note: most biopsies do not contain tumors, but changes occur in these samples, nevertheless),
- develop an RNA strategy (now licensed to a company),
- build large tissue microarrays for protein prognosticators,
- develop methylation assays (see below).
Program 4: DNA methylation biomarkers for cancer progression.
Background. For prostate cancer, the sources of materials for prognostics include needle biopsies, blood, and urine, as well as prostatectomies. Prognostic tests can include pathology, DNA, RNA, or protein, or combinations thereof. DNA is more stable than RNA and is found in all of these types of samples. The methylation profile is unaltered in partially degraded DNA after cell death.
Objective. Develop a cancer prognosticator based on DNA rather than RNA or protein.
Approaches. We developed a method that samples hundreds of thousands of DNA methylation loci throughout the genome. The prostate tumor samples in Program 3 are being screened to find differences in DNA methylation that are prognostic. The markers are subsequently converted to an assay platform that can be used in a clinical service laboratory.