RESEARCH OVERVIEW
Our goal is to understand how microbes within communities adapt to environmental change. This is not an easy task. On the one hand, microbial communities consist of a large number of different species resulting in a complex interaction network that is constantly changing over time. 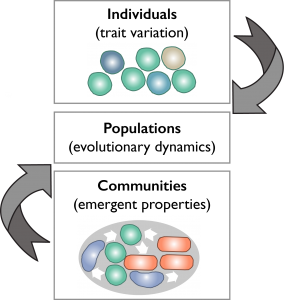 On the other hand, with recent advances in single-cell techniques, we are only now beginning to identify and characterise microbial individual’s traits relevant for adaptation. In our lab we use an interdisciplinary approach to understand how individual cell variation influence evolutionary dynamics, and how community dynamics in turn influence evolutionary outcomes, i.e. eco-evolutionary feedback loops.
On the other hand, with recent advances in single-cell techniques, we are only now beginning to identify and characterise microbial individual’s traits relevant for adaptation. In our lab we use an interdisciplinary approach to understand how individual cell variation influence evolutionary dynamics, and how community dynamics in turn influence evolutionary outcomes, i.e. eco-evolutionary feedback loops.
Our general working approach is to build mathematical models to formulate specific predictions. We then test these predictions by doing experiments in highly controlled environments. The overall expectation from these experiments is to generate general concepts that extend beyond specific bacterial species and communities.
- Rodríguez-Verdugo A. 2021. Evolving interactions and emergent functions in microbial consortia. Early-Career Special Series, mSystems. 6: e00774-21 https://doi.org/10.1128/mSystems.00774-21
RESEARCH LINES
I) MICROBIAL EVOLUTION IN A COMMUNITY CONTEXT. We study how species interactions give rise to emergent properties at the community level that feedback to influence the evolution of populations and how this evolution affects the stability and functionality of microbial communities.
Three major questions that drive our research are:
- How do higher-order interactions and indirect effects shape the evolution of multi-species systems?
- How do species interactions evolve and give rise to new community functionalities; e.g. novel collective metabolism?
- What type of species interactions lead to community-level cohesion?
To address these questions, we use a bottom-up approach. That is, we use communities of reduced complexity – synthetic communities– to measure and quantify species interactionsin highly controlled environments. Once we have a mechanistic understanding on species interactions, we experimentally expose these synthetic communities to a desired condition for hundreds of generations and we use whole-genome sequencing to see what mutations accumulated in each population at the end of the experiment.
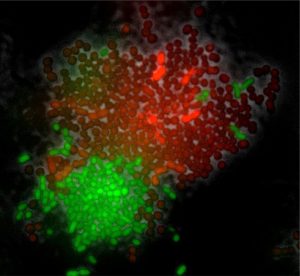
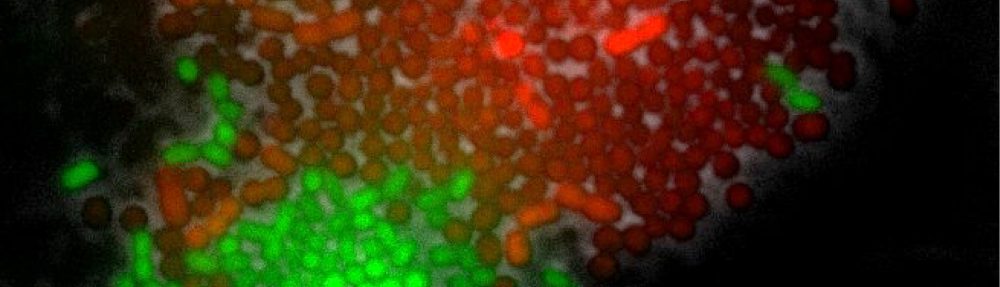
Fluorescence image of Acinetobacter johnsonii (in red; mCherry) interacting with Pseudomonas putida (in green; GFP). Credit: Alejandra Rodriguez Verdugo, ETH Zurich.
We are studying the interaction between Acinetobacter johnsonii and Pseudomonas putida, two bacterial species that were originally isolated from a polluted aquifer in Denmark. This consortium is relevant for bioremediation given that species collectively degrade toxic and recalcitrant compounds that pollute the environment. We also like to work with this system because species and their interactions are easy to manipulate; e.g. we can switch species interactions from competitive to exploitative depending on the carbon source provided.
Relevant publications:
- Al-Tameemi, Z., and A. Rodríguez-Verdugo. 2024. Microbial diversification is maintained in an experimentally evolved synthetic community. mSystems. e01053-24 https://doi.org/10.1128/msystems.01053-24
- Rodríguez-Verdugo A. and M. Ackermann. 2021. Rapid evolution destabilizes species interactions in a fluctuating environment. The ISME Journal. 15:450-460.
https://doi.org/10.1038/s41396-020-00787-9 - Rodríguez-Verdugo A., C. Vulin and M. Ackermann. 2019. The rate of environmental fluctuations shapes ecological dynamics in a two-species microbial system. Ecology Letters. 22:838-846 https://doi.org/10.1111/ele.13241
II) MICROBIAL RESPONSES TO CLIMATE CHANGE. Microbial communities perform pivotal functions such the decomposition of plant litter. This important ecosystem function emerges from the interaction of multiple species working as a collective. A key question is: how do collective functions could persist, in the long-term, in face of environmental stressors such as global warming?
We have joined a collaboration, The Loma Ridge Climate Change Experiment, with UCI professors Steve Allison, Kathleen Treseder, Jennifer Martiny and Adam Martiny aiming to understand how the plant litter microbiome respond to drought and heat in Southern California. We have assembled multispecies consortia representative of the leaf litter microbiome from the Loma Ridge grassland site to understand how response to drought and heat affect community-level functions important for ecosystems functioning.
Relevant publications:
- Abdoli, P., C. Vulin, M. Lepiz, A.B. Chase, C. Weihe, A. Rodríguez-Verdugo. 2023. Substrate complexity buffers negative interactions in a synthetic microbial community of leaf litter degraders. FEMS Microbiology Ecology. 100(8) fiae102 https://doi.org/10.1093/femsec/fiae102
- Martiny J.B.H., A.C. Martiny, E. Brodie, A.B. Chase, A. Rodríguez-Verdugo, K.K. Treseder, S. Allison. 2023. Investigating the eco-evolutionary response of microbiomes to environmental change. Ecology Letters. https://doi.org/10.1111/ele.14209
III) ANTIBIOTIC RESISTANCE IN AN ENVIRONMENTAL CONTEXT. We study the evolution of antibiotic resistance in the context of environmental stressors (e.g., thermal stress) and species interactions.
Two major questions that drive our research are:
- How do bacteria respond to combinations of stresses such as heat and antibiotics?
- How do species interactions influence the evolution of antibiotic resistance?
We have established two exciting collaborations to explore these questions. The first collaboration with UCLA professors Pamela Yeh and Van Savage, focuses on the link between heat stress adaptation and the evolution of antibiotic resistance in E.coli. The second collaboration with Mexican researchers at Cinvestav consists of evolving bacterial strains from Cuatro Ciénegas, Coahuila, in Mexico, under different antibiotics to study the evolution of resistance in natural communities. Check this external link to learn more about Cuatro Ciénegas, Coahuila
Relevant publications:
- Bullivant A., N. Lozano-Huntelman, K. Tabibian, V. Leung, D. Armstring, H. Dudley, V.M. Savage, A. Rodríguez-Verdugo, P.J. Yeh. 2024. Evolution under thermal stress affects Escherichia coli‘s resistance to antibiotics. bioRxiv. https://doi.org/10.1101/2024.02.27.582334.
- Cruz-Loya M., E. Tekin, T. Manzhu Kang, N. Cardona, N. Lozano-Hunterlman, A. Rodríguez-Verdugo, V.M. Savage, P.J. Yeh. 2021. Antibiotics shifts the temperature response curve of Escherichia coli growth. mSystems. 6:e00228-21 https://doi.org/10.1128/mSystems.00228-21
- Rodríguez-Verdugo A., N. Lozano-Huntelman, M. Cruz-Loya, V. Savage and P. Yeh. 2020. Compounding effects of climate warming and antibiotic resistance. iScience. 23, 24 April 2020, 101024 https://doi.org/10.1016/j.isci.2020.101024