Overview and goals
Our lab explores the innate immune response pathways that organisms use to defend themselves against viral pathogens. We answer fundamental mechanistic questions regarding the enzymatic synthesis, specific recognition, and multi-level regulation of nucleotide-based second messengers in diverse immunological settings. Cyclic nucleotide signaling molecules are responsible for relaying and amplifying signals in nearly every biological context and have recently emerged as critical players in specialized antiviral pathways in animals and in microbial antiphage defense systems. To investigate diverse immune systems and their antiviral signaling molecules, we use a combination of recombinant protein expression, biochemical and chemical analysis, enzymology, as well as modern protein structural approaches (X-ray crystallography & cryo-EM).
Our research in this rapidly developing field will identify novel second messengers and reveal uncharacterized antiviral signaling pathways controlled by these compounds. Recently, many surprising mechanistic links between eukaryotic and prokaryotic immune signaling have been found including the discovery that bacterial genomes harbor genes encoding structural and functional homologs of critical protein components of the animal innate immune system. Through exploration of the ancient connections between prokaryotic and eukaryotic immunity we can transform our understanding of our own immune system and reveal new avenues for exploitation of these pathways for medicinal purposes and as foundation to develop new biotechnological tools.
Cyclic nucleotide signaling
A major focus of the lab is to understand the activation mechanisms and function of enzymes in immune pathways that generate specialized nucleotide signaling molecules. Recently, we provided the first evidence that cyclic CMP and cyclic UMP are important signals involved in a bacterial immune response to phage infection, produced through the action of enzymes in a defense system called Pycsar (pyrimidine cyclase system for antiphage resistance). We also revealed that a defense system known as CBASS (cyclic oligonucleotide-based antiphage signaling system), which functions through production of cyclic dinucleotide and trinucleotide second messengers, has evolutionary ties to the human cGAS-STING antiviral signaling pathway. In both Pycsar and CBASS, an enzyme is activated by phage to produce a cyclic nucleotide signal that then binds to a receptor that in-turn facilitates the destruction of the host cell to prevent the virus from completing its replication cycle. Through our research on these newly described defense systems, we will identify novel enzymatic activities and cyclic nucleotide products, clarify the mechanistic details underlying enzyme activation by phage infection, and define the rules that govern nucleotide selection at the level of the enzyme (substrate) and receptor (ligand).
TIR domains and NADase enzymology
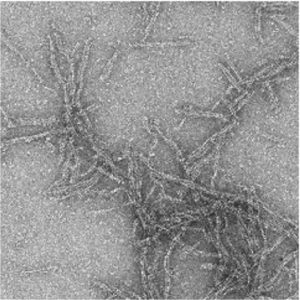
Other nucleotide-derived signaling molecules also play important functions in innate immunity, especially those compounds generated by enzymes that degrade the ubiquitous and critical metabolite, NAD+ (β-nicotinamide adenine dinucleotide). NAD+glycosyl hydrolases of the TIR domain family (Toll/Interleukin-1 receptor) rapidly degrade NAD+ in response to stimuli such as a viral infection or neuronal injury and some even go a step further to generate novel cyclized products. TIR enzymes have been found in many of these newly defined bacterial immune defense systems (including Pycsar & CBASS) where they function to deplete NAD+ in the cell and cause cellular growth arrest or cell death under certain conditions. TIR enzymes require protein-protein contacts brought about through homo-oligomerization for efficient NAD+ turnover and in many cases these complexes can grow quite large in vitro (tens to hundreds of copies). We will explore the dynamic growth properties of these TIR oligomers, determine the molecular switches that control NADase activation (through ligand recognition), and investigate the consequences of NAD+degradation product signaling on host-cell survival and viral restriction.
TIR domain oligomeric filaments observed via cryo-EM
“Orphan” immune systems
Bacteria have evolved an amazing variety of protective measures in order to survive under constant threat of infection by the bacteriophage that vastly outnumber them. Bioinformatics analysis has recently revealed the existence of hundreds of distinct predicted immune defense systems encoded within bacterial genomes. Excitingly, many of these predicted systems contain protein domains with no biochemically confirmed functions and in many instances no known three-dimensional structures. With these predictions as a guide, we will begin to functionally characterize new immunological systems and identify novel mechanisms of defense while also experimentally determining structures of new protein folds and domains. We will use structure-guided phylogenetic analysis to assess patterns of conservation of these systems amongst diverse organisms. Surprisingly, the human genome also encodes many uncharacterized proteins with predicted or known structural homology to proteins involved in immune signaling. We will pursue the elucidation of the functional roles of these “orphan” enzymes and receptors to establish new possible therapeutic targets in the treatment of disease and to expand our overall basic understanding of human physiology and cell signaling.