Investigating the METABOLIC INDEX In-vivo
Investigator: [Dr. Jenu Chacko now at the University of Wisconsin]
‘Nanoscopy’ based method gives us a magnified view of the nanometer scale objects and their interactions that happen inside the cells. At this size scale, molecules and their interactions play a crucial role in identifying the cause and effect observed. Many Nanoscopy methods arose in the last few decades to visualize these small dimensions in live cells. Methods such as image correlation techniques/diffusion studies unveil information details on the size, shape, environment, partner molecules, interaction type, interaction distances, etc. The Digman lab addresses to enhance the currently available imaging techniques in order to address more parameters connected with protein-protein interactions pointing towards a macro scale explanation of live animal studies. As a team, we study the autofluorescence of live cells and tissues in order to answer the current questions arising in the biomedical and clinical fields. The redox state of the cell through NADH fluorescence lifetime imaging, protein interactions with cellular auto-fluorescence and in-vivo deep tissue imaging and tracking of fluorescent and auto-fluorescent species, etc. are some of the undergoing projects.
FLIM and FCS Analysis of the DNA Damage Response
Investigators: Murata, M., Kong, X., & Yokomori, K.
This research project utilizes fluorescence lifetime imaging microscopy (FLIM) and fluorescence correlation spectroscopy (FCS) to study the dynamics of DNA repair protein recruitment to damage sites and transcriptional regulation of DNA repair proteins. First, by using FLIM to gather high spatiotemporal information about the redox state of NADH, we are able to study the cellular metabolic response to different types of DNA damage caused by gamma irradiation, bleomycin, hydrogen peroxide, and laser-induced damage as well as the transcriptional activity of NADH in the nucleus during DNA damage repair.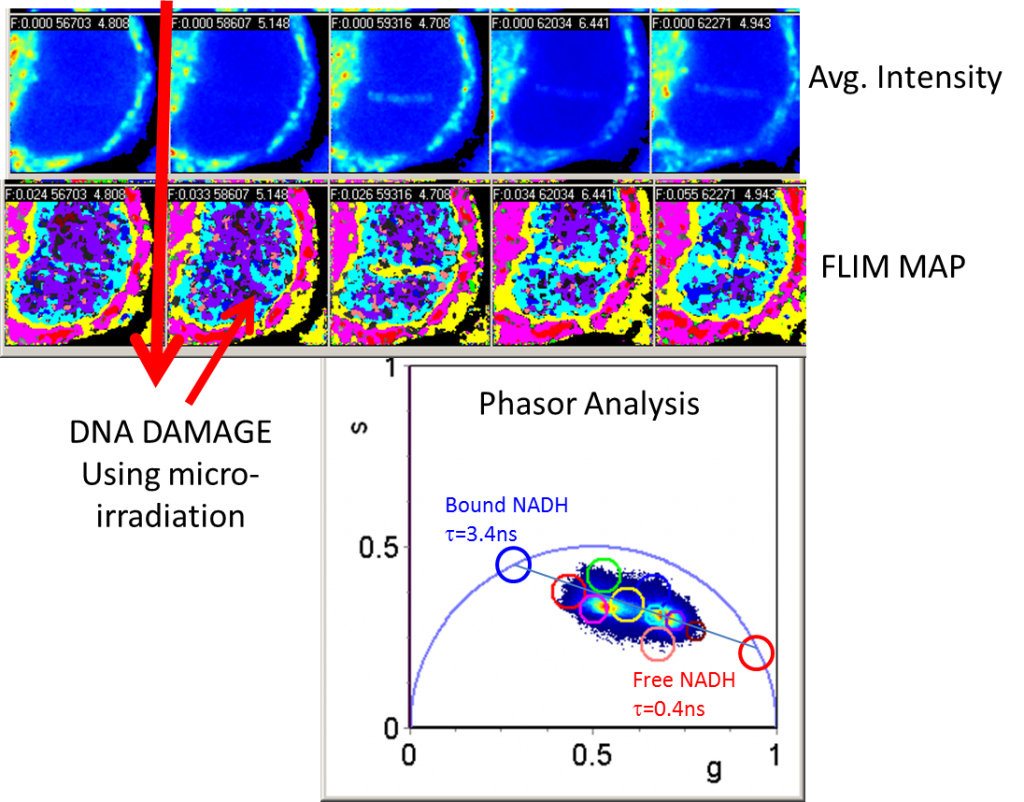
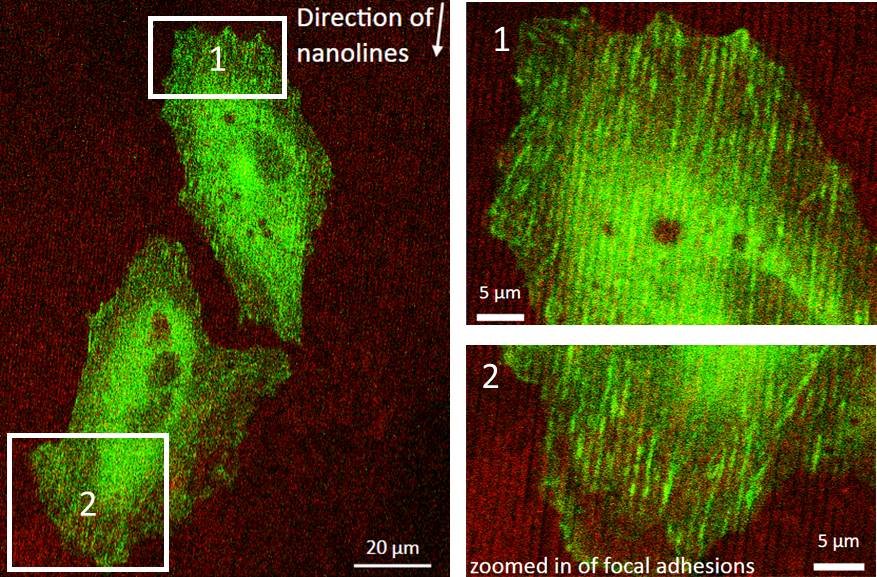
Cell migration and focal adhesion formation on nanopatterned surfaces
Investigator: Emma Mah
Reference: Liang, E.I., Mah,E.J., Yee A.F., and Digman, M.A. Integr. Biol., 201 (2017),9, 145-155, DOI: 10.1039/C6IB00193A,
Studying cell-to-substrate or cell-to-cell interactions can provide important insight to the understanding of cell functions, mechanics, and signaling. Cell signaling is regulated by the interactions of membrane bound proteins with the extra cellular matrix (ECM). Organization and composition of components within the ECM changes cell migration,adhesion, and proliferation patterns. These changes are often regulated by focal adhesion (FA) complexes, which are composed of over 150 proteins.
Understanding these cell-to-substrate interactions will provide information to optimize materials used to promote wound healing and increase biocompatibility of implants. I am to develop a platform to study the interaction of cells on nanopatterns subtracts in vitro. In addition, I would like to used these patterns to study the interactions of tumor associated macrophages and breast cancer cells.
Tumor associated macrophages (TAMs) are known to act as “leader cells” for guiding breast cancer cells onto collagen fibers to blood vessels and initialize metastasize. The collagen provides a physical path in which the TAM can be directed to the vessels. Thus, the use of nanolines will provide a path for migration to occur. Current studies are done in vivo which ignore for possible factors which may affect their observations (e.g signaling from other cells, foreign chemoattactants). Thus, using this in vitro method simplifies the system and allows control of any factors which may contribute to the interaction between TAMs and cancer cells. A variety of imaging and analysis techniques (z-phasors, tICS, FLIM) can be used which will allow me to visualize real-time protein dynamics in cells and quantify their movements.
Metabolic mapping of different cell lines via 2-Photon fluorescence lifetime imaging of the coenzyme NADH
Investigator: Ning Ma
Biochemical estimation of NADH concentration is a useful method for monitoring cellular metabolism, because the NADH/NAD+ reduction-oxidation pair is crucial for electron transfer in the mitochondrial electron chain. Our aim is to using two-photon fluorescence lifetime imaging (FLIM) to derive functional maps of intracellular reduction-oxidation ration in vivo via measurement of the fluorescence lifetimes and the ratio of free and protein-bound NADH.
Non-invasive live imaging of stem cell signature metabolic states
Investigator: Ning Ma
To model human development we need to recapitulate developmental processes by mimicking growth factor production from a source and monitor metabolic responses to GFs on a cell-by-cell basis. To do this we propose to test whether stimulation of growth factor pathways, including the WNT and BMP pathway,alters the metabolic output of hESCs and to study metabolism in living pluripotent cells both in vitro and in vivo.
Mitchondria transport and metabolic shifts within Invadopodia
Investigator: Emma Mah
Cancer is the second leading cause of death in the world. Invadopodia are actin rich  protrusions in which cancer cells form to invade and degrade the surrounding extracellular matrix (ECM). Eventually, the cell is able migrate into other parts of the body and continue their grow (e.g. metastasis). These protrusions are known to favor formation on stiffer substrates and their function is regulated based on the metabolism on the cell. Stiffer matrices promote increased actin polymerization and focal adhesion stabilization which are energy demanding processes within the cell. Their has been recent discovery that within the protrusions, oxidative phosphorylation is the main method of metabolism and mitochondria are recruited to these sites. Mitochondria are key organelles which produce energy in the cell (i.e. ATP) and are know travel towards the leading edge of the cell during migration. However, there is little knowledge of whether mitochondria recruitment also play a key role in invadopodia formation and function and if this is dependent on matrix stiffness. We use 2-photon excitation of NADH and fluorescent lifetime imaging microscopy (FLIM) to determine metabolic indexes of the cell on various stiffness of collagen and tracking to visualize mitochondria transport towards the leading edge. Thus, we aim to bridge the mechanosensing and metabolism pathways within invadopodia. This will allow us to determine if invadopodia formation on stiffer substrates also correlate with mitochondria, shifts metabolic pathways, and matrix degradation.
protrusions in which cancer cells form to invade and degrade the surrounding extracellular matrix (ECM). Eventually, the cell is able migrate into other parts of the body and continue their grow (e.g. metastasis). These protrusions are known to favor formation on stiffer substrates and their function is regulated based on the metabolism on the cell. Stiffer matrices promote increased actin polymerization and focal adhesion stabilization which are energy demanding processes within the cell. Their has been recent discovery that within the protrusions, oxidative phosphorylation is the main method of metabolism and mitochondria are recruited to these sites. Mitochondria are key organelles which produce energy in the cell (i.e. ATP) and are know travel towards the leading edge of the cell during migration. However, there is little knowledge of whether mitochondria recruitment also play a key role in invadopodia formation and function and if this is dependent on matrix stiffness. We use 2-photon excitation of NADH and fluorescent lifetime imaging microscopy (FLIM) to determine metabolic indexes of the cell on various stiffness of collagen and tracking to visualize mitochondria transport towards the leading edge. Thus, we aim to bridge the mechanosensing and metabolism pathways within invadopodia. This will allow us to determine if invadopodia formation on stiffer substrates also correlate with mitochondria, shifts metabolic pathways, and matrix degradation.
Spatio-temporal Mapping of the Estrogen Receptor
Investigator: Ramon Rocca
Estrogen receptors (ERs), specifically ERα, are crucial proteins in breast and ovarian cancer. ERs are over-exp ressed in around 70% of breast cancer cases, referred to as “ER-positive”. Either binding of estrogen to the ER stimulates proliferation of mammary cells, with the resulting increase in cell division and DNA replication, leading to mutations or estrogen metabolism produces genotoxic waste. In both cases, the result is disruption of cell cycle, DNA repair and apoptosis that lead to tumor formation[1].
ressed in around 70% of breast cancer cases, referred to as “ER-positive”. Either binding of estrogen to the ER stimulates proliferation of mammary cells, with the resulting increase in cell division and DNA replication, leading to mutations or estrogen metabolism produces genotoxic waste. In both cases, the result is disruption of cell cycle, DNA repair and apoptosis that lead to tumor formation[1].
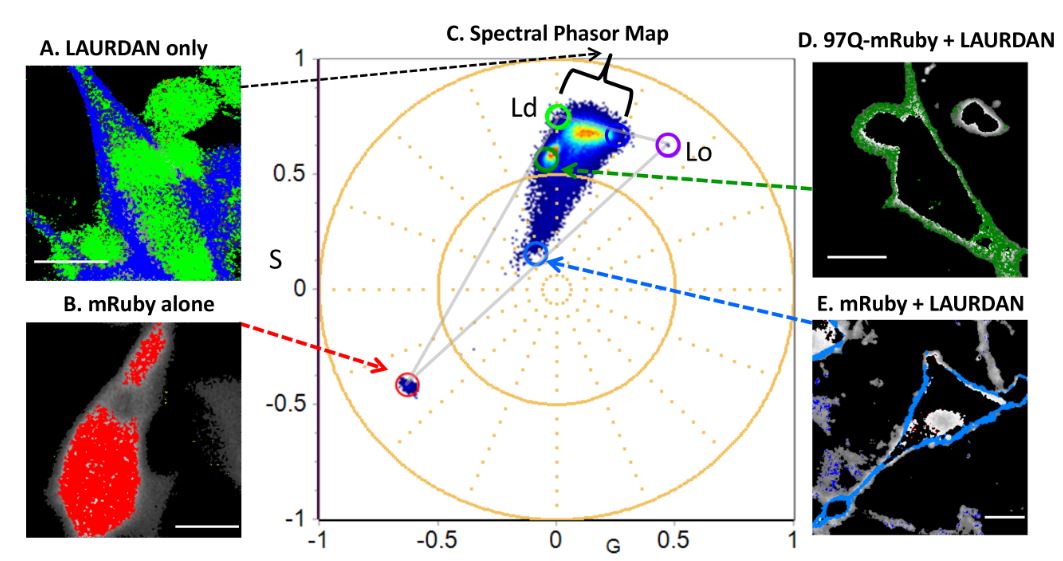
The phasor-FLIM fingerprints reveal shifts from OXPHOS to enhanced glycolysis in Huntington Disease; Investigator: Sara Sameni; Reference: Sameni, S., Malacrida, L., Tan, Z. & Digman, M. A. Alteration in Fluidity of Cell Plasma Membrane in Huntington Disease Revealed by Spectral Phasor Analysis. Scientific Reports 8, 734, doi:10.1038/s41598-018-19160-0 (2018).
Figure below: Mapping the emission spectra of LAURDAN (liquid ordered (Lo) and disordered (Ld) membranes) and mRuby in combination using spectral Phasor plot in differentiated PC12 cells expressing control,mRuby only, LAURDAN only and 97PolyQ Htt.
Phasor approach to examine and characterize metabolic dysfunction in neurodegenerative diseases; Investigator: Sara Sameni;Reference: Sameni et al, Scientific Reports 6, Article number: 34755 (2016); doi:10.1038/srep34755
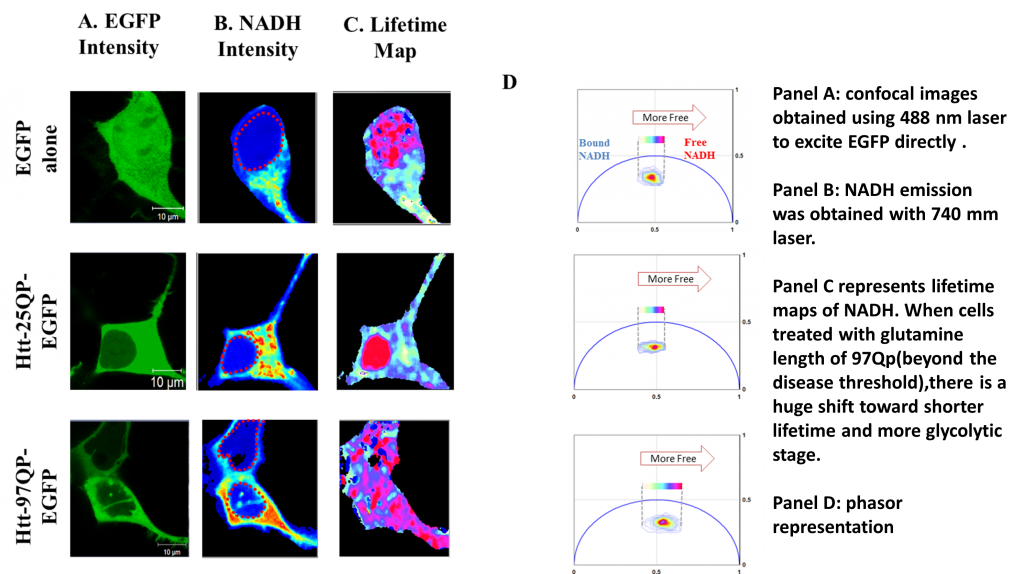 Protein aggregation and inclusion in affected brain regions, is the hallmark of many neurodegenerative diseases including Huntington disease. Better understanding of the cellular mechanism disturbed by mutation, and in particular metabolic dysfunction that has shown to be a co-exiting condition, can lead to more effective therapeutic outcome. My effort for such analysis is to measure variation in NADH to NAD associated with protein over expression and aggregation in living cell using a very precise and sensitive approach, phasor florescence lifetime imaging microscopy. In addition, I will use number and brightness (N&B) method to measure the molecular brightness of the protein aggregates non-invasively in entire cell to further characterize the aggregates using a label free approach, non-invasively.
Protein aggregation and inclusion in affected brain regions, is the hallmark of many neurodegenerative diseases including Huntington disease. Better understanding of the cellular mechanism disturbed by mutation, and in particular metabolic dysfunction that has shown to be a co-exiting condition, can lead to more effective therapeutic outcome. My effort for such analysis is to measure variation in NADH to NAD associated with protein over expression and aggregation in living cell using a very precise and sensitive approach, phasor florescence lifetime imaging microscopy. In addition, I will use number and brightness (N&B) method to measure the molecular brightness of the protein aggregates non-invasively in entire cell to further characterize the aggregates using a label free approach, non-invasively.